GenRA Manual: Web Application
Previous chapter: Overview & Background
GenRA aims to mirror the steps undertaken in a typical analogue or category approach (Patlewicz et al., 2017). Source analogues are identified using chemical or bioactivity fingerprints or a combination of both. From there, an evaluation of the data gaps is performed before predictions are made for all toxicity (activity) effects of interest. The main steps of the analogue/category workflow as implemented in GenRA are shown in Figure 1. The default use case in GenRA is to make binary predictions of in vivo toxicity taken from ToxRefDB but it is also possible to make in vivo potency toxicity estimates and binary hitcall predictions of ToxCast assays.
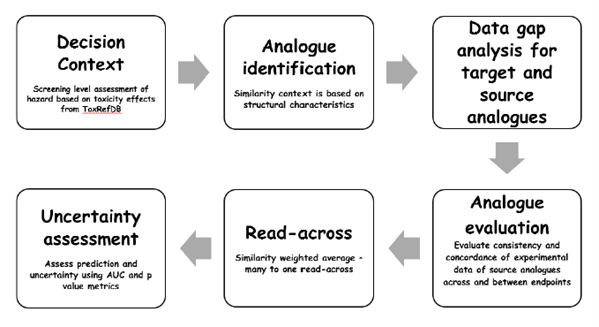
Entry Points for Using GenRA
The starting point for GenRA assumes the user has a target chemical of interest for which data is lacking. This can be introduced into GenRA in one of several ways:
- If the user is already using the EPA CompTox Chemicals Dashboard (CCD) for a specific substance of interest, a hyperlink to access the GenRA may be found on the left-hand panel of the chemical landing page (Figure 2).
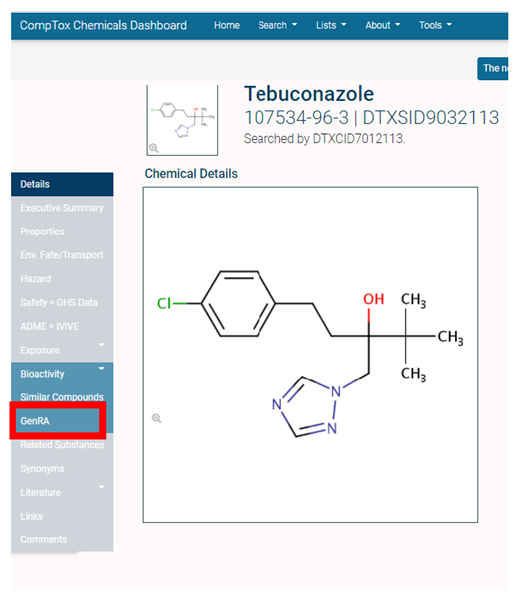
Figure 2. EPA CompTox Chemicals Dashboard - GenRA link may also be found under the Tools menu on the main CCD home page. This will redirect the user to the main home page of GenRA at https://comptox.epa.gov/genra/ (Figure 3).
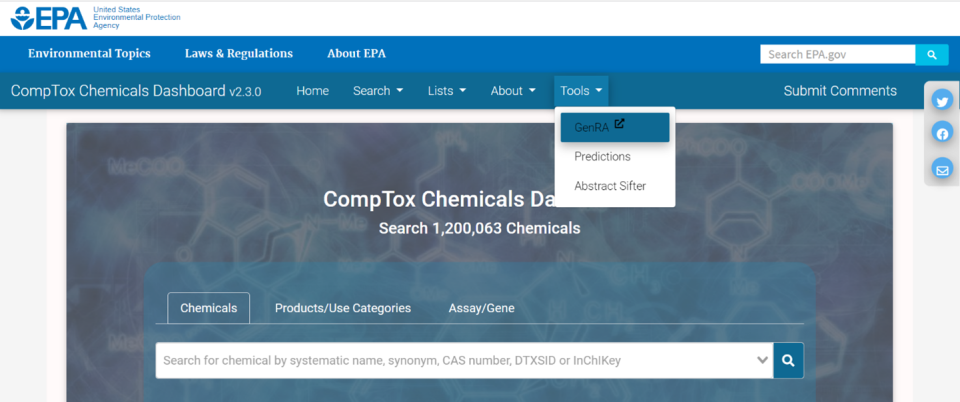
Next chapter: Selecting a Chemical for Study
